The R profiler
The R profiler is an amazing way to find where in your code (or someone else's code) lies some inefficiency. For example, the profiler helped in this challenge on the R wiki. See also the profiling section on the http://cran.r-project.org/doc/manuals/R-exts.html#Profiling-R-code-for-speed document
What is wrong with it
The profiler, however, is not able to trace uses of loops (for, while or repeat), and consequently will not identify loops as the cause of ineffiency of the code. This is a shame, because loops in R are usually closely related to inefficiency. For example, if we profile this code:
1 Rprof( )
2 x <- numeric( )
3 for( i in 1:10000){
4 x <- c( x, rnorm(10) )
5 }
6 Rprof( NULL )
7 print( summaryRprof( ) )
8
we get :
$ time Rscript script1.R
$by.self
self.time self.pct total.time total.pct
"rnorm" 0.16 100 0.16 100
$by.total
total.time total.pct self.time self.pct
"rnorm" 0.16 100 0.16 100
$sampling.time
[1] 0.16
real 0m7.043s
user 0m5.170s
sys 0m0.675s
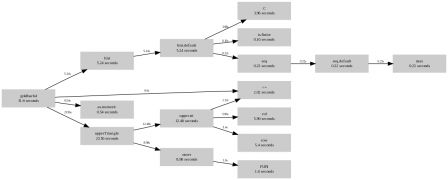
So the profiler only reports about 0.22 seconds, when the actual time taken is more about 5 seconds. We can show that by wrapping the entire for loop in a function:
1 Rprof( )
2 ffor <- function(){
3 x <- numeric( )
4 for( i in 1:10000){
5 x <- c( x, rnorm(10) )
6 }
7 }
8 ffor()
9 Rprof( NULL )
10 print( summaryRprof( ) )
11
which gives this :
$ time Rscript script2.R
$by.self
self.time self.pct total.time total.pct
"ffor" 5.14 96.3 5.34 100.0
"rnorm" 0.20 3.7 0.20 3.7
$by.total
total.time total.pct self.time self.pct
"ffor" 5.34 100.0 5.14 96.3
"rnorm" 0.20 3.7 0.20 3.7
$sampling.time
[1] 5.34
real 0m6.434s
user 0m5.151s
sys 0m0.698s
The ffor function takes 100 pourcent of the times, and rnorm takes only 3.7 percent of the time, instead of 100 percent, which would be the conclusion of the first example.
But in real life, it is not possible to wrap every loop in a function as this will massively break a lot of code. Instead, we could make the profiler aware of loops. This is the purpose of the patch I posted to R-devel
The details of the implementation
The patch actually only takes place in (several places of) the file eval.c
In the do_for function, a context is created for the "for" loop, using the begincontext function:
1033 begincontext(&cntxt, CTXT_LOOP, R_NilValue, rho, R_BaseEnv, R_NilValue,
1034 mkChar("[for]") );
The change here appears on the second line and simply adds a bit of information to the context that is created, similar changes are also made on the functions do_repeat and do_while.
Next, we need to grab this information at each tick of the profiler, which is the job of the doprof function:
168 if ((cptr->callflag & (CTXT_FUNCTION | CTXT_BUILTIN))
169 && TYPEOF(cptr->call) == LANGSXP) {
170 SEXP fun = CAR(cptr->call);
171 if (!newline) newline = 1;
172 fprintf(R_ProfileOutfile, "\"%s\" ",
173 TYPEOF(fun) == SYMSXP ? CHAR(PRINTNAME(fun)) :
174 "<Anonymous>");
175 } else if( (cptr->callflag & CTXT_LOOP) ){
176 if (!newline) newline = 1;
177 fprintf(R_ProfileOutfile, "\"%s\" ", CHAR(cptr->callfun) );
178 }
The else branch will be executed when the context is a a loop context, and we just retrieve the callfun string we created in the do_for function.
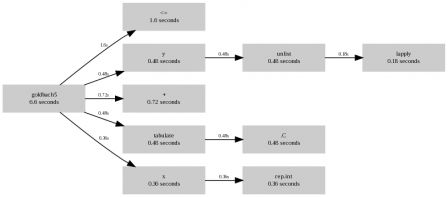
Now, with this R patched, and compiled, Rprof is able to record loops
[]$ /home/romain/workspace/R-trunk/bin/Rscript script1.R
$by.self
self.time self.pct total.time total.pct
"[for]" 5.28 97.4 5.42 100.0
"rnorm" 0.14 2.6 0.14 2.6
$by.total
total.time total.pct self.time self.pct
"[for]" 5.42 100.0 5.28 97.4
"rnorm" 0.14 2.6 0.14 2.6
$sampling.time
[1] 5.42
[]$ head Rprof.out
sample.interval=20000
"[for]"
"[for]"
"[for]"
"rnorm" "[for]"
"[for]"
"[for]"
"[for]"
"[for]"
"[for]"

 Future plans for this package contain:
Future plans for this package contain: