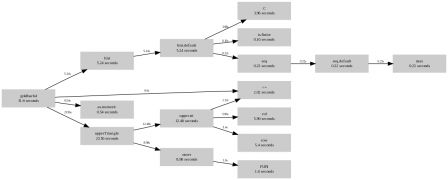
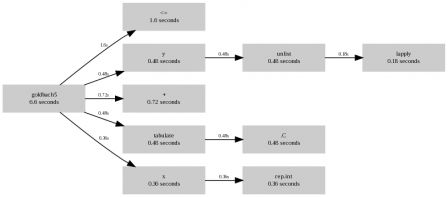
Following this post, there is still room for improvement. Recall the last implementation (goldbach5)
12 goldbach5 <- function(n) { 13 xx <- 1 : n 14 xx <- xx[isprime(xx) > 0][-1] 15 16 # generates row indices 17 x <- function(N){ 18 rep.int( 1:N, N:1) 19 } 20 # generates column indices 21 y <- function(N){ 22 unlist( lapply( 1:N, seq.int, to = N ) ) 23 } 24 z <- xx[ x(length(xx)) ] + xx[ y(length(xx)) ] 25 z <- z[z <= n] 26 tz <- tabulate(z, n ) 27 tz[ tz != 0 ] 28 } 29
The first thing to notice right away is that when we build xx in the first place, we are building integers from 1 to n, and check if they are prime afterwards. In that case, we are only going to need odd numbers in xx, so we can build them dircetly as:
31 xx <- seq.int( 3, n, by = 2) 32 xx <- xx[isprime(xx) > 0]
The next thing is that, even though the goldbach5 version only builds the upper triangle of the matrix, which saves some memory, we don't really need all the numbers, since eventually we just want to count how many times each of them appears. For that, there is no need to store them all in memory.
The version goldbach6 below takes this idea forward implementing the counting in C using the .C interface. See this section of writing R extensions for details of the .C interface.
30 goldbach6 <- function( n ){ 31 xx <- seq.int( 3, n, by = 2) 32 xx <- xx[isprime(xx) > 0] 33 out <- integer( n ) 34 tz <- .C( "goldbach", xx =as.integer(xx), nx = length(xx), 35 out = out, n = as.integer(n), DUP=FALSE )$out 36 tz[ tz != 0 ] 37 } 38
and the C function that goes with it
1 #include <R.h> 2 3 void goldbach(int * xx, int* nx, int* out, int* n){ 4 int i,j,k; 5 6 for( i=0; i<*nx; i++){ 7 for( j=i; j<*nx; j++){ 8 k = xx[i] + xx[j] ; 9 if( k > *n){ 10 break; 11 } 12 out[k-1]++ ; 13 } 14 } 15 } 16
We need to build the shared object
$ R CMD SHLIB goldbach.c gcc -std=gnu99 -I/usr/local/lib/R/include -I/usr/local/include -fpic -g -O2 -c goldbach.c -o goldbach.o gcc -std=gnu99 -shared -L/usr/local/lib -o goldbach.so goldbach.o -L/usr/local/lib/R/lib -lR
and load it in R:
> dyn.load( "goldbach.so" )
And now, let's see if it was worth the effort
> system.time( out <- goldbach6(100000) ) user system elapsed 0.204 0.005 0.228 > system.time( out <- goldbach5(100000) ) user system elapsed 4.839 2.630 7.981 > system.time( out <- goldbach4(100000) ) user system elapsed 28.425 5.932 38.380
We could also have a look at the memoty footprint of each of the three functions using the memory profiler.
> gc(); Rprof( "goldbach4.out", memory.profiling=T ); out <- goldbach4(100000); Rprof(NULL)
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 150096 4.1 350000 9.4 350000 9.4
Vcells 213046 1.7 104145663 794.6 199361285 1521.1
> gc(); Rprof( "goldbach5.out", memory.profiling=T ); out <- goldbach5(100000); Rprof(NULL)
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 150093 4.1 350000 9.4 350000 9.4
Vcells 213043 1.7 162727615 1241.6 199361285 1521.1
> gc(); Rprof( "goldbach6.out", memory.profiling=T ); out <- goldbach6(100000); Rprof(NULL)
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 150093 4.1 350000 9.4 350000 9.4
Vcells 213043 1.7 130182091 993.3 199361285 1521.1
> rbind( summaryRprof( filename="goldbach4.out", memory="both" )$by.total[1,] ,
+ summaryRprof( filename="goldbach5.out", memory="both" )$by.total[1,],
+ summaryRprof( filename="goldbach6.out", memory="both" )$by.total[1,] )
total.time total.pct mem.total self.time self.pct
"goldbach4" 32.08 100 712.6 1.26 3.9
"goldbach5" 6.66 100 306.7 2.80 42.0
"goldbach6" 0.22 100 0.2 0.00 0.0